Segmentation is a crucial step of 3D rendering. In P3D this task is done in the lower section of the Input tab.
The aim of segmentation is to find the boundary of a tissue structure, which can be transformed into a virtual 3D object. In general, segmentation is a process which is difficult to fully automate. The best results are obtained if the structure is clearly separated from the remainder by a substantial image contrast, and if prior anatomical knowledge is used to guide the segmentation process.
P3D offers several general segmentation methods for finding the object contours. The contour finding process can further be restricted by an enclosing volume-of-interest (VOI). The Smooth box allows enabling a smoothing function during the segmentation. Note, however, that smoothing will change the volume of the segments and is thus not recommended for small objects.
Each Input study has its own segmentation definition which will be updated when switching between the studies. Therefore, to begin with a segmentation task, first select the appropriate Input study, and then adjust the the segmentation method.
ITK-based Implementations
Some of the segmentation methods are based on the use of the ITK (Insight Toolkit) libraries. ITK can be used under the open-source BSD license which allows unrestricted use, including use in commercial products (see www.itk.org). The ITK-based methods are clearly denoted with ITK in brackets.
Note: PMOD Technologies cannot be held liable for permanent support of the ITK interface, nor for the performance of the provided libraries.
Region Growing Methods
"Region growing algorithms have proven to be an effective approach for image segmentation. The basic approach of a region growing algorithm is to start from a seed region (typically one or more pixels) that are considered to be inside the object to be segmented. The pixels neighboring this region are evaluated to determine if they should also be considered part of the object. If so, they are added to the region and the process continues as long as new pixels are added to the region. Region growing algorithms vary depending on the criteria used to decide whether a pixel should be included in the region or not, the type of connectivity used to determine neighbors, and the strategy used to visit neighboring pixels." The ITK Software Guide.
Segmentation Preview
As a help during the definition of the segmentation criterion a preview of the expected result is shown in the Input display if the Ovr (Overlay) box is checked. The example below illustrates REGION GROWING after clicking into a mouse kidney. Note how the color of the segmentation overlay changes depending on the color table used to display the images.
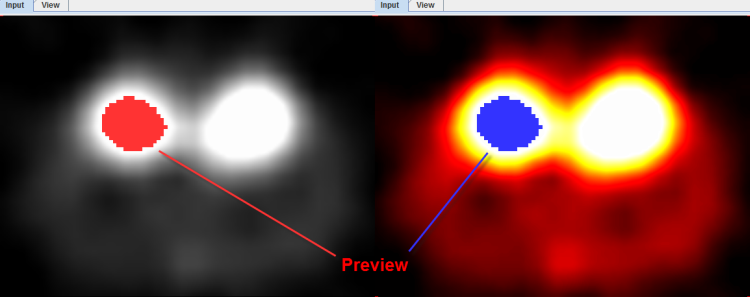
To start with segmentation, navigate to a slice (in any direction) which shows the tissue of interest and click with the left mouse button onto a central point. Then adjust the criterion until the overlay indicates a promising segmentation.
Please use the ![]() toggle button in the taskbar for enlarging/shrinking the area of the segmentation preview image. Note that the image display controls are not accessible in the enlarged mode, so the image presentation should be adjusted beforehand.
toggle button in the taskbar for enlarging/shrinking the area of the segmentation preview image. Note that the image display controls are not accessible in the enlarged mode, so the image presentation should be adjusted beforehand.
For further expanding of the 2D view port activate the Hide segmentation controls  button located under the segmentation settings. Note that upon activation of this button the Segmentation panel collapse and only the segmentation method selection list is still available. Therefore, any further settings should be set beforehad.
button located under the segmentation settings. Note that upon activation of this button the Segmentation panel collapse and only the segmentation method selection list is still available. Therefore, any further settings should be set beforehad.
CAUTION: The preview only shows the segmentation within the current slice. As there may exist indirect connections through neighboring slices, additional pixels may be contained in the final segment.